This protocol describes the use of NEIGHBOR (see Fig. 6.3.1), included in the PHYLIP 3.6 package, which is distributed by Joe Felsenstein (University of Washington) and is one of the most widely used software packages in phylogeny studies. NEIGHBOR is the PHYLIP implementation of Neighbor Joining (Saitou and Nei, 1987). Distance estimation is performed using DNADIST or PROTDIST (Support Protocols 1 and 2). To accomplish the bootstrap procedure, first resample the sites using SEQBOOT (Support Protocol 3), then apply DNADIST or PROTDIST, run NEIGHBOR, and extract the bootstrap tree using CONSENSE (Support Protocol 3). Finally, the resulting tree can be drawn using a program such as TreeView (UNIT 6.2) or NJplot (Perriere and Gouy, 1996).
Necessary Resources
PHYLIP, which stands for 'The PHYLogeny Inference Package, is a collection of applications that were developed to help advanced computer users such as genetics engineers or scientists in related. PHYLIP is a package of programs for inferring phylogenies. The algorithm is a fairly simple adaptation of the one used in the program SOKAL, which was formerly in this package and has been superseded by MIX. It requires two passes through each tree to count the numbers of reversions. An R interface for PHYLIP. Contribute to liamrevell/Rphylip development by creating an account on GitHub. And maintain their software on GitHub — the largest and most advanced development platform in the world. Sign up for free Dismiss. And package builds for the project 'Rphylip: An R interface for PHYLIP.' An R interface for.
Hardware
PHYLIP executables are available for pre-386 DOS, 386/486/Pentium DOS, Windows 3.1, Windows 95/98/NT, 68k Macintosh, or PowerMac. The PHYLIP C source code is also available for Unix, Linux, or VMS systems.
Software
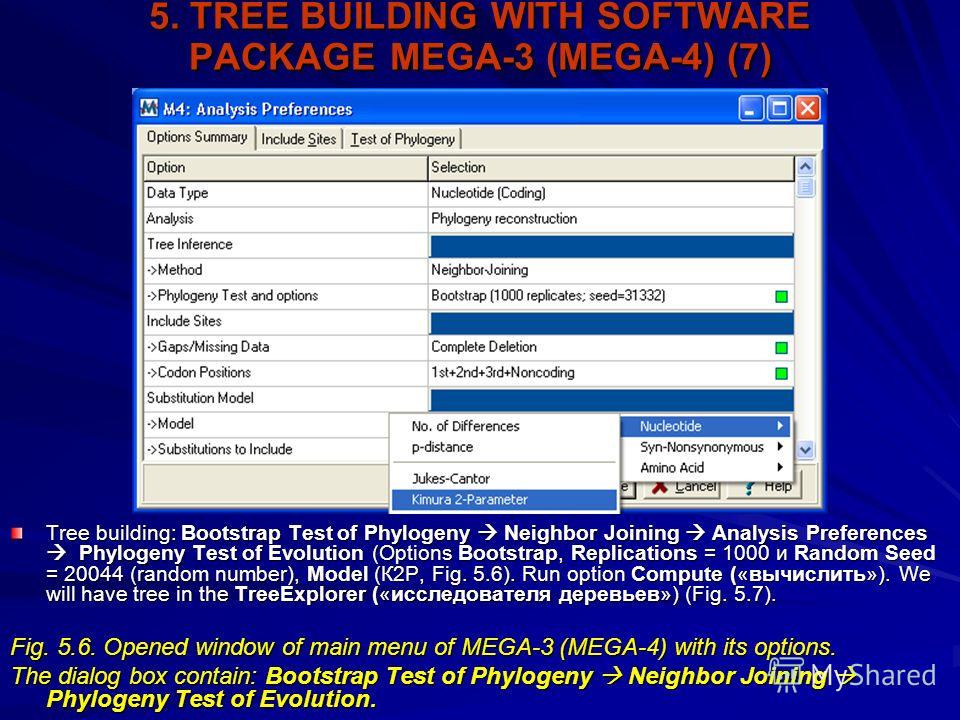
Phylip Format
PHYLIP is available for free from
http://evolution.Qenetics.washinQton.edu/phylip. html. The package contains C source codes, documentation files, and a number of different types of executables. Its Web page contains information on PHYLIP and ways to transfer the executables, source code, and documentation. The documentation is remarkably clear and complete, and provides a number of useful references.
Files
NEIGHBOR requires a distance matrix (or a set of distances matrices when the bootstrap procedure is used), which is estimated by DNADIST (Support Protocol 1) or PROTDIST (Support Protocol 2) from a multiple sequence alignment. The file contains a number of taxa on its first line. Each taxon starts a new line with the taxon name, followed by the distance to the other taxa, and there is a new line after every nine distances. Taxon names have ten characters and must be blank-filled to be of that length. The default matrix format is square (Fig. 6.3.2) with zero distances on the diagonal. In the case of multiple matrices, as obtained with the bootstrap, matrices are given in the same format one after the other, without omitting the number of taxa at the beginning of each new matrix.
1. Download and install PHYLIP according to the program documentation (see Necessary Resources, above).
2. Generate a distance matrix for the multiple sequence alignment of interest by running either DNADIST (for DNA sequence alignments; see Support Protocol 1) or PROTDIST (for protein sequence alignments; see Support Protocol 2).
3. Begin a NEIGHBOR session in PHYLIP by double clicking on its icon.
4. At the prompt, enter the distance matrix file name and the name for the outfile, which will contain a simple representation of the output tree. The default files are infile and outfile, respectively, but the authors strongly recommend redefining these files to avoid possible confusions or deleting previously computed files.
Phylip Package Software
When a file called infile already exists in the PHYLIP directory, NEIGHBOR does not ask for the input file and reads the existing infile. Similarly, the option of renaming the output is only given if a file called outfile already exists. If no such file exists, NEIGHBOR automatically writes the output to a file called outfile.
5. Once done, the user has to select among numerous options (see Fig. 6.3.3), which, a priori, have to be used with their default values, except M in the case of the bootstrap procedure. When options have been determined, type 'Y' to run NEIGHBOR.
These options are as follows. N defines the method to be used; NJ (default option) has to be preferred over UPGMA, which assumes a molecular clock. Bellawood installation guide. O makes it possible to specify which species is to be used to root the tree; when O is on, the user is asked for the rank of the outgroup species in the input (matrix) file, otherwise the default outgroup species is the first; this outgroup (rooting) species is used in the tree printed in the outfile. L and R have to be switched on when the matrix is not square but lower-triangular and upper-triangular, respectively. S has to be on when the data contain subreplicates; it allows NEIGHBOR to read the input data, but the number of replicates is ignored. J enables one to choose a random order of species; the user is then asked for a 'seed'; however, NEIGHBOR is almost insensitive to species ordering. M has to be used in the case of the bootstrap procedure (Support Protocol 3) to provide the number of pseudo-matrices. 0 defines the terminal type; this may affect the ability of the programs to display their menus and results, but the 'none' option is usually satisfying. The 1 and 2 options are used to check the data and the progress of run; the authors suggest switching them off, notably for large trees and bootstrap studies. When 3 is Yes (default value), the tree or trees are printed in the outfile; this is useful to quickly visualize trees with moderate numbers of taxa, in case of unique data set. When 4 is Yes (default value), the trees are written in Newick format in the outtree file, and can then be drawn using TreeView (UNIT 6.2) or, in case of multiple data sets, combined by CONSENSE to obtain the bootstrap tree (Support Protocol 3). To change the default values, simply type the option character. For example, typing 2 changes the progress of run status from Yes to No, and typing 2 again returns one to Yes.
6. Finally, NEIGHBOR asks for the outtree file, which will contain the tree in Newick format (UNIT 6.2). The resulting tree can be visualized in the outfile, but a better view is obtained by applying TreeView (UNIT 6.2) to the outtree file.
The option of renaming the outtree file is only given if a file called outtree already exists. If no such file exists, NEIGHBOR automatically writes the output to a file called outtree, which may be a source of confusion. Inferred trees are unrooted and written in Newick format (UNIT 6.2). For example, the BIONJ tree in Figure 6.3.4 is made of three subtrees, containing (Candida_tr, Candida_al, and Saccharomy), (Taphrina_d and Protomyces) and (Athelia_bo, Spongipell, and Filobasidi), respectively, as can be shown from its TreeView representation (Fig. 6.3.5; see UNIT 6.2 for discussion of TreeView and Newick). Each subtree is made up of two subtrees or taxa; the numbers in Figure 6.3.4 indicate the branch lengths. Both trees in Figure 6.3.4 have identical topologies (even when the way they are encoded in Newick format looks quite different) but (slightly) different branch lengths.
Applying NEIGHBOR to the matrix of Figure 6.3.2, one obtains in the outfile the tree shown in Figure 6.3.6, while in the outtree file we have the second tree from Figure 6.3.4, in Newick format. This tree is equivalent to that of Figure 6.3.5.
7. To assess the tree quality, bootstrap the tree according to Support Protocol 3.
From Current Protocols in Bioinformatics Online Copyright © 2002 John Wiley & Sons, Inc. All rights reserved.
CURRENT PROTOCOLS IN BIOINFORMATICS CHAPTER 6 INFERRING EVOLUTIONARY RELATIONSHIPS UNIT 6.3 Getting a Tree Fast: Neighbor Joining and Distance-Based Methods
SUPPORT PROTOCOL 1: DISTANCE MATRIX ESTIMATION FROM DNA (OR RNA) SEQUENCES USING DNADIST
SUPPORT PROTOCOL 1: DISTANCE MATRIX ESTIMATION FROM DNA (OR RNA) SEQUENCES USING DNADIST
Wwe 2k15 full game download for pc. WWE 2K15 free. download full Version for PC Game setup with a single and direct download link. Download WWE 2K15 and play on your own computer or laptop. WWE 2K15 Overview WWE 2K15 was developed by Yuke's, Visual Concepts and n-Space (mobile version) and it was Published by 2K Sports. WWE 2K15 PC Download is a professional fighting video game developed by Yuke's and Visual Concepts and offers 2K Sports for PlayStation 3 (PS3), PlayStation 4 (PS4)., Xbox 360, Xbox One and Windows PC. It is successor of WWE 2K16. Since the days of THQ and Jacks Pacific, the WWE or WWF if you're from that generation, has always been a subject for video games. Whether it was WWE Smackdown!Here Comes The Pain, Smackdown! Shut Your Mouth or Smackdown vs Raw, there was always a licensed game on hand to let you play as your spandex-wearing heroes. While the licence has changed hands to 2K, the same holds true in.
Distance estimation is the first step in reconstructing a phylogenetic tree using a distance-based method. DNADIST, from the PHYLIP package, estimates the pairwise evolutionary distances between nucleotide sequences under various models of nucleotide substitutions. These models account for hidden substitutions and incorporate knowledge about the mutation process. Distance estimation is based on the maximum-likelihood principle (Swofford et al., 1996). The model choice is sensitive and influences the distance values, and then the tree to be constructed. DNADIST reads a multiple sequence alignment and outputs a distance matrix. When the bootstrap procedure is used, the input file contains the pseudo-alignments one after the other, and the output file contains the corresponding pseudo-matrices in the same order.
Necessary Resources
Hardware
PHYLIP executables are available for pre-386 DOS, 386/486/Pentium DOS, Windows 3.1, Windows 95/98/NT, 68k Macintosh, or PowerMac. The PHYLIP C source code is also available for Unix, Linux, or VMS systems.
Software
DNADIST is part of the PHYLIP package. PHYLIP is available for free from http://evolution.penetics.washinpton.edu/phvlip.html. The package contains C source codes, documentation files, and a number of different types of executables. Its Web page contains information on PHYLIP and ways to transfer the executables, source code, and documentation. The documentation is remarkably clear and complete, and provides a number of useful references.
Files
DNADIST requires DNA multiple sequence alignments in PHYLIP format, as obtained from alignment programs such as ClustalX (UNIT
2.3). The first line contains the number of taxa and sites; next come the taxon data with a new line per taxon. Taxon names have ten characters and must be blank-filled to be of that length. The taxon names are followed by the sequences, which must either be 'interleaved' or 'sequential' (Figs. 6.3.7 and 6.3.8). The sequences can have internal blanks in the sequence but there must be no extra blanks at the end of the terminated line. The three symbols N, X and ? indicate an unknown nucleotide while a dash (-) indicates a deletion. In the case of multiple data sets, as provided by SEQBOOT, pseudo-alignments are given in the same format one after the other, without omitting the number of taxa and the number of sites at the beginning of each new set.
1. Download and install the PHYLIP package and, initialize a DNADIST session by double clicking on its icon.
2. At the prompt, enter the sequence alignment file name and the name for the output, which will contain the distance matrix. The default files are infile and outfile, respectively, but the authors strongly recommend redefining these files to avoid possible confusion, or deletion of previously computed files.

Cyberlink powerdvd 16 activation url. When a file called infile already exists in the PHYLIP directory, DNADIST does not ask for the input file and reads the existing infile. Similarly, the option of renaming the output is only given if a file called outfile already exists. If no such file exists, DNADIST automatically writes the output to a file called outfile.
3. Then the menu of Figure 6.3.9 appears, which asks for important and sensitive choices.
The remaining steps of this protocol primarily describe options requiring in-depth explanations or where the default values have often to be changed. More details are given in the DNADIST documentation. To change the default values, simply type the option character. For example, typing 'I' changes the sequence format from interleaved to sequential, and typing 'I' again returns to the interleaved format.
Set the parameters
4. D defines the substitution model. All models assume that sites evolve independently. The four available models are nested, i.e., Jukes-Cantor is a special case of Kimura, which is a special case of F84, which is a special case of LogDet. Jukes-Cantor (Jukes and Cantor, 1969) assumes only one substitution rate, Kimura (Kimura, 1980) allows for a difference between transition and transversion rates, while F84 (Kishino and Hasegawa, 1989; Felsenstein and Churchill, 1996) is similar to
Kimura but allows for different frequencies of the four nucleotides, and LogDet does not impose any restriction on the 16 rates (except those induced by the Markovian nature of the process). So LogDet (Steel, 1994) is the most flexible model, but is often overparametrized, unless the sequences are very long (say >3000). F84 (the default option) is a good compromise, notably when the base frequencies are not equal. When they are almost equal, Kimura is a good choice, while Jukes-Cantor is overly simple in most cases.
Note that all sites (informative or not) must be given to DNADIST for these models to be used in the correct way.
5. G asks whether or not the substitution rates vary across sites. Biologically speaking, the answer is clearly yes. It has been demonstrated that the Gamma distribution (Swofford et al., 1996), which is defined by a parameter usually denoted as a, is a good model to account for this variability. a was estimated between 0.05 and 1.0 for numerous data sets (Yang, 1996), which indicates that rates strongly vary across sites (variability increases as a decreases). However, the default option of DNADIST is to not correct for this variability (i.e., a = ¥), which is a common practice.
Jin and Nei (1990) recommend using a = 1.0 or 2.0. The authors of this unit have recently demonstrated (Guindon and Gascuel, 2002) that uncorrected distances are often better suited, especially when the molecular clock is more or less satisfied. Therefore, a pragmatic approach is to use the default option, and to check whether or not using a reasonable value (e.g., 1.0 or 2.0) for a changes the result. A software program to estimate the most appropriate value of a is also available via the authors' Web page (http://www.lirmm.fr/~w3ifa/MAAS/).
However, DNADIST does not use the standard a parameter, but rather the 'coefficient of variation' (CV), which is equal to 1/a2. One obtains CV = 4.0, 1.0, and 0.25, when a = 0.5, 1.0 and 2.0, respectively. Moreover, the LogDet model cannot be combined with the gamma correction.
6. T asks for the transition/transversion ratio. The default value is 2.0, and there is no way to estimate this value within PHYLIP.
Hopefully, the results are not very sensitive to the value of this parameter (unless it is extreme). It is possible to estimate it using simple formulas from Kimura (1980).
Phylip Format
PHYLIP is available for free from
http://evolution.Qenetics.washinQton.edu/phylip. html. The package contains C source codes, documentation files, and a number of different types of executables. Its Web page contains information on PHYLIP and ways to transfer the executables, source code, and documentation. The documentation is remarkably clear and complete, and provides a number of useful references.
Files
NEIGHBOR requires a distance matrix (or a set of distances matrices when the bootstrap procedure is used), which is estimated by DNADIST (Support Protocol 1) or PROTDIST (Support Protocol 2) from a multiple sequence alignment. The file contains a number of taxa on its first line. Each taxon starts a new line with the taxon name, followed by the distance to the other taxa, and there is a new line after every nine distances. Taxon names have ten characters and must be blank-filled to be of that length. The default matrix format is square (Fig. 6.3.2) with zero distances on the diagonal. In the case of multiple matrices, as obtained with the bootstrap, matrices are given in the same format one after the other, without omitting the number of taxa at the beginning of each new matrix.
1. Download and install PHYLIP according to the program documentation (see Necessary Resources, above).
2. Generate a distance matrix for the multiple sequence alignment of interest by running either DNADIST (for DNA sequence alignments; see Support Protocol 1) or PROTDIST (for protein sequence alignments; see Support Protocol 2).
3. Begin a NEIGHBOR session in PHYLIP by double clicking on its icon.
4. At the prompt, enter the distance matrix file name and the name for the outfile, which will contain a simple representation of the output tree. The default files are infile and outfile, respectively, but the authors strongly recommend redefining these files to avoid possible confusions or deleting previously computed files.
Phylip Package Software
When a file called infile already exists in the PHYLIP directory, NEIGHBOR does not ask for the input file and reads the existing infile. Similarly, the option of renaming the output is only given if a file called outfile already exists. If no such file exists, NEIGHBOR automatically writes the output to a file called outfile.
5. Once done, the user has to select among numerous options (see Fig. 6.3.3), which, a priori, have to be used with their default values, except M in the case of the bootstrap procedure. When options have been determined, type 'Y' to run NEIGHBOR.
These options are as follows. N defines the method to be used; NJ (default option) has to be preferred over UPGMA, which assumes a molecular clock. Bellawood installation guide. O makes it possible to specify which species is to be used to root the tree; when O is on, the user is asked for the rank of the outgroup species in the input (matrix) file, otherwise the default outgroup species is the first; this outgroup (rooting) species is used in the tree printed in the outfile. L and R have to be switched on when the matrix is not square but lower-triangular and upper-triangular, respectively. S has to be on when the data contain subreplicates; it allows NEIGHBOR to read the input data, but the number of replicates is ignored. J enables one to choose a random order of species; the user is then asked for a 'seed'; however, NEIGHBOR is almost insensitive to species ordering. M has to be used in the case of the bootstrap procedure (Support Protocol 3) to provide the number of pseudo-matrices. 0 defines the terminal type; this may affect the ability of the programs to display their menus and results, but the 'none' option is usually satisfying. The 1 and 2 options are used to check the data and the progress of run; the authors suggest switching them off, notably for large trees and bootstrap studies. When 3 is Yes (default value), the tree or trees are printed in the outfile; this is useful to quickly visualize trees with moderate numbers of taxa, in case of unique data set. When 4 is Yes (default value), the trees are written in Newick format in the outtree file, and can then be drawn using TreeView (UNIT 6.2) or, in case of multiple data sets, combined by CONSENSE to obtain the bootstrap tree (Support Protocol 3). To change the default values, simply type the option character. For example, typing 2 changes the progress of run status from Yes to No, and typing 2 again returns one to Yes.
6. Finally, NEIGHBOR asks for the outtree file, which will contain the tree in Newick format (UNIT 6.2). The resulting tree can be visualized in the outfile, but a better view is obtained by applying TreeView (UNIT 6.2) to the outtree file.
The option of renaming the outtree file is only given if a file called outtree already exists. If no such file exists, NEIGHBOR automatically writes the output to a file called outtree, which may be a source of confusion. Inferred trees are unrooted and written in Newick format (UNIT 6.2). For example, the BIONJ tree in Figure 6.3.4 is made of three subtrees, containing (Candida_tr, Candida_al, and Saccharomy), (Taphrina_d and Protomyces) and (Athelia_bo, Spongipell, and Filobasidi), respectively, as can be shown from its TreeView representation (Fig. 6.3.5; see UNIT 6.2 for discussion of TreeView and Newick). Each subtree is made up of two subtrees or taxa; the numbers in Figure 6.3.4 indicate the branch lengths. Both trees in Figure 6.3.4 have identical topologies (even when the way they are encoded in Newick format looks quite different) but (slightly) different branch lengths.
Applying NEIGHBOR to the matrix of Figure 6.3.2, one obtains in the outfile the tree shown in Figure 6.3.6, while in the outtree file we have the second tree from Figure 6.3.4, in Newick format. This tree is equivalent to that of Figure 6.3.5.
7. To assess the tree quality, bootstrap the tree according to Support Protocol 3.
From Current Protocols in Bioinformatics Online Copyright © 2002 John Wiley & Sons, Inc. All rights reserved.
CURRENT PROTOCOLS IN BIOINFORMATICS CHAPTER 6 INFERRING EVOLUTIONARY RELATIONSHIPS UNIT 6.3 Getting a Tree Fast: Neighbor Joining and Distance-Based Methods
SUPPORT PROTOCOL 1: DISTANCE MATRIX ESTIMATION FROM DNA (OR RNA) SEQUENCES USING DNADIST
SUPPORT PROTOCOL 1: DISTANCE MATRIX ESTIMATION FROM DNA (OR RNA) SEQUENCES USING DNADIST
Wwe 2k15 full game download for pc. WWE 2K15 free. download full Version for PC Game setup with a single and direct download link. Download WWE 2K15 and play on your own computer or laptop. WWE 2K15 Overview WWE 2K15 was developed by Yuke's, Visual Concepts and n-Space (mobile version) and it was Published by 2K Sports. WWE 2K15 PC Download is a professional fighting video game developed by Yuke's and Visual Concepts and offers 2K Sports for PlayStation 3 (PS3), PlayStation 4 (PS4)., Xbox 360, Xbox One and Windows PC. It is successor of WWE 2K16. Since the days of THQ and Jacks Pacific, the WWE or WWF if you're from that generation, has always been a subject for video games. Whether it was WWE Smackdown!Here Comes The Pain, Smackdown! Shut Your Mouth or Smackdown vs Raw, there was always a licensed game on hand to let you play as your spandex-wearing heroes. While the licence has changed hands to 2K, the same holds true in.
Distance estimation is the first step in reconstructing a phylogenetic tree using a distance-based method. DNADIST, from the PHYLIP package, estimates the pairwise evolutionary distances between nucleotide sequences under various models of nucleotide substitutions. These models account for hidden substitutions and incorporate knowledge about the mutation process. Distance estimation is based on the maximum-likelihood principle (Swofford et al., 1996). The model choice is sensitive and influences the distance values, and then the tree to be constructed. DNADIST reads a multiple sequence alignment and outputs a distance matrix. When the bootstrap procedure is used, the input file contains the pseudo-alignments one after the other, and the output file contains the corresponding pseudo-matrices in the same order.
Necessary Resources
Hardware
PHYLIP executables are available for pre-386 DOS, 386/486/Pentium DOS, Windows 3.1, Windows 95/98/NT, 68k Macintosh, or PowerMac. The PHYLIP C source code is also available for Unix, Linux, or VMS systems.
Software
DNADIST is part of the PHYLIP package. PHYLIP is available for free from http://evolution.penetics.washinpton.edu/phvlip.html. The package contains C source codes, documentation files, and a number of different types of executables. Its Web page contains information on PHYLIP and ways to transfer the executables, source code, and documentation. The documentation is remarkably clear and complete, and provides a number of useful references.
Files
DNADIST requires DNA multiple sequence alignments in PHYLIP format, as obtained from alignment programs such as ClustalX (UNIT
2.3). The first line contains the number of taxa and sites; next come the taxon data with a new line per taxon. Taxon names have ten characters and must be blank-filled to be of that length. The taxon names are followed by the sequences, which must either be 'interleaved' or 'sequential' (Figs. 6.3.7 and 6.3.8). The sequences can have internal blanks in the sequence but there must be no extra blanks at the end of the terminated line. The three symbols N, X and ? indicate an unknown nucleotide while a dash (-) indicates a deletion. In the case of multiple data sets, as provided by SEQBOOT, pseudo-alignments are given in the same format one after the other, without omitting the number of taxa and the number of sites at the beginning of each new set.
1. Download and install the PHYLIP package and, initialize a DNADIST session by double clicking on its icon.
2. At the prompt, enter the sequence alignment file name and the name for the output, which will contain the distance matrix. The default files are infile and outfile, respectively, but the authors strongly recommend redefining these files to avoid possible confusion, or deletion of previously computed files.
Cyberlink powerdvd 16 activation url. When a file called infile already exists in the PHYLIP directory, DNADIST does not ask for the input file and reads the existing infile. Similarly, the option of renaming the output is only given if a file called outfile already exists. If no such file exists, DNADIST automatically writes the output to a file called outfile.
3. Then the menu of Figure 6.3.9 appears, which asks for important and sensitive choices.
The remaining steps of this protocol primarily describe options requiring in-depth explanations or where the default values have often to be changed. More details are given in the DNADIST documentation. To change the default values, simply type the option character. For example, typing 'I' changes the sequence format from interleaved to sequential, and typing 'I' again returns to the interleaved format.
Set the parameters
4. D defines the substitution model. All models assume that sites evolve independently. The four available models are nested, i.e., Jukes-Cantor is a special case of Kimura, which is a special case of F84, which is a special case of LogDet. Jukes-Cantor (Jukes and Cantor, 1969) assumes only one substitution rate, Kimura (Kimura, 1980) allows for a difference between transition and transversion rates, while F84 (Kishino and Hasegawa, 1989; Felsenstein and Churchill, 1996) is similar to
Kimura but allows for different frequencies of the four nucleotides, and LogDet does not impose any restriction on the 16 rates (except those induced by the Markovian nature of the process). So LogDet (Steel, 1994) is the most flexible model, but is often overparametrized, unless the sequences are very long (say >3000). F84 (the default option) is a good compromise, notably when the base frequencies are not equal. When they are almost equal, Kimura is a good choice, while Jukes-Cantor is overly simple in most cases.
Note that all sites (informative or not) must be given to DNADIST for these models to be used in the correct way.
5. G asks whether or not the substitution rates vary across sites. Biologically speaking, the answer is clearly yes. It has been demonstrated that the Gamma distribution (Swofford et al., 1996), which is defined by a parameter usually denoted as a, is a good model to account for this variability. a was estimated between 0.05 and 1.0 for numerous data sets (Yang, 1996), which indicates that rates strongly vary across sites (variability increases as a decreases). However, the default option of DNADIST is to not correct for this variability (i.e., a = ¥), which is a common practice.
Jin and Nei (1990) recommend using a = 1.0 or 2.0. The authors of this unit have recently demonstrated (Guindon and Gascuel, 2002) that uncorrected distances are often better suited, especially when the molecular clock is more or less satisfied. Therefore, a pragmatic approach is to use the default option, and to check whether or not using a reasonable value (e.g., 1.0 or 2.0) for a changes the result. A software program to estimate the most appropriate value of a is also available via the authors' Web page (http://www.lirmm.fr/~w3ifa/MAAS/).
However, DNADIST does not use the standard a parameter, but rather the 'coefficient of variation' (CV), which is equal to 1/a2. One obtains CV = 4.0, 1.0, and 0.25, when a = 0.5, 1.0 and 2.0, respectively. Moreover, the LogDet model cannot be combined with the gamma correction.
6. T asks for the transition/transversion ratio. The default value is 2.0, and there is no way to estimate this value within PHYLIP.
Hopefully, the results are not very sensitive to the value of this parameter (unless it is extreme). It is possible to estimate it using simple formulas from Kimura (1980).
7. C allows user-defined categories, for example to specify that third-position bases have a different rate than first and second positions. This option allows the user to make up to 9 categories of sites, but, as for the LogDet model, using too many categories can make the model overparametrized. The user is asked for the relative rates within each category. The assignment of rates to sites is then made by reading a file whose default name is 'categories.'
An example and more details are given in the DNADIST documentation. There is no program from PHYLIP for estimating the different rates, but just as for the above ratio these parameters are not very sensitive (unless extreme).
8. W allows to select subsets of sites. Basically it has to remain 'No' (the default value), unless the user wants to check the influence of various categories of sites.
See DNADIST documentation for more details.
9. F must remain as Yes in any practical situation.
10. L defines the matrix format, square (default value) or lower-triangular.
11. M has to be used in the bootstrap procedure (see Support Protocol 3). The user is then asked for the number of pseudo-alignments in the input file. Otherwise the default value (No) is required.
12. I defines the multiple sequence alignment format, which is interleaved or sequential (Fig. 6.3.7 and 6.3.8, respectively).
13. Once all options have been determined, type 'Y' to compute the distance matrix.
With the working example of Figure 6.3.7 and all default values, DNADIST returns the matrix of Figure 6.3.2.
From Current Protocols in Bioinformatics Online Copyright © 2002 John Wiley & Sons, Inc. All rights reserved.
CURRENT PROTOCOLS IN BIOINFORMATICS CHAPTER 6 INFERRING EVOLUTIONARY RELATIONSHIPS UNIT 6.3 Getting a Tree Fast: Neighbor Joining and Distance-Based Methods
SUPPORT PROTOCOL 2: DISTANCE MATRIX ESTIMATION FROM PROTEINS USING PROTDIST
Was this article helpful?
| Biosignal Processing and Analysis This lab focuses on using, analysing and processing EEG data and provides a platform for EEG data analysis and visualization, to understand the correlations of neural activity through electroencephalography data. The lab is an education platform for engineers and biologists without major requirements for learning methods in signal processing. |
| Bioinformatics and Data Science in Biotechnology This lab is a connection of bioinformatics experiments performed using R programming. Educating this will allow users to learn how to use R as an open source language for learning bioinformatics data processing. Specifically, this lab will help analyse biological sequence data using simple R code snippets. Writing and Reading Sequence Data in R || Pairwise sequence alignment of Protein or DNA sequences || Guanine-Cytosine Content Analysis and Basics of DNA Sequence Statistics || Reading FASTA using SequinR |
| Neurophysiology Virtual Lab (pilot) Neurophysiology is the study of nervous system function. Primarily, it is connected with neurobiology, psychology, neurology, clinical neurophysiology, electrophysiology, biophysical neurophysiology, ethology, neuroanatomy, cognitive science and other brain sciences. Brain Slice Preparation || Simple Neuron Model - the HH neuron || Patch Clamp Technique || Current Clamp Technique || Voltage Clamp Technique || Study of Synaptic Transmission (Remote trigger) || Measuring Field Potentials Using MEA chips || Understanding the Passive Properties of a Simple Neuron (Remote trigger) || Effects of Ion Channels in Membrane Biophysics (Remote trigger) || Effect of Noise on Spiking Neurons (Remote trigger) |
| Neuron Simulation Virtual Lab (pilot) This lab uses a graphical web-based Neuron simulator and models a section of excitable neuronal membrane using the Hodgkin-Huxley equations. Various experiments will deal with the several parameters of Hodgkin-Huxley equations and will model resting and action potentials, voltage and current clamp, pharmacological effects of drugs that block specific channels etc. This lab complements some of the exercises in the Virtual Neurophysiology lab. Modeling resting potentials in Neurons || Modeling action potentials || Modeling the delayed rectifier Potassium channels || Modeling the sodium ion channel and its effects on neural signaling || Current Clamp protocol || Voltage Clamp Protocol || Understanding Frequency-Current relationship || Understanding first spike latency - current relationship || Voltage-Current (VI) plot || Effects of pharmacological blockers on action potential |
| Biochemistry Virtual Lab I Biochemistry is the study of the chemical processes in living organisms. It deals with the structures and functions of cellular components such as proteins, carbohydrates, lipids, nucleic acids and other biomolecules. The experiments included in Biochemistry Virtual Lab I are fundamental in nature, dealing with the identification and classification of various carbohydrates, acid-base titrations of amino acids, isolation of proteins from their natural sources, etc. Qualitative Analysis of Carbohydrates || Isoelectric Precipitation of Proteins: Casein from Milk || Quantitative Estimation of Amino Acids by Ninhydrin || Separation of Amino Acids by Thin Layer Chromatography || Estimation of Saponification Value of Fats/Oils. || Detection of Adulteration in Milk || Qualitative Analysis of Amino Acid || Estimation of Iodine Value of Fats and Oils || Titration Curves of Aminoacids || Estimation of blood glucose by Glucose oxidase method |
| Biochemistry Virtual Lab II Biochemistry Virtual Lab II deals with topics like enzymology, purification of plant pigments and natural products as well as estimation of iodine value and saponification value of fats and oils. Isolation of β -Amylase from Sweet Potato || Gelatin Zymography || Extraction of Caffeine from Tea || Construction of Maltose Standard Curve by DNS Method || Isolation of Plant Pigments by Column Chromatography || Structural Studies of Phycobiliproteins from Spirulina || Construction of Protein Standard Curve using Folin's Lowry Method || Effect of Substrate Concentration on Enzyme Kinetics || Effect of temperature on enzyme kinetics || Hydrolysis of Ester using orange peel esterase |
| Population ecology Virtual Lab I A population is a collection of individuals of the same species that live together in a region. Population ecology is the study of populations (especially population abundance) and how they change over time. Crucial to this study are the various interactions between a population and its resources. Studies on simple models of interacting species is the main focus this simulation oriented lab. Lab I focuses on introduction of principles of population ecology for UG/PG students. Population with Continuous and Discrete Growth || Spread of a Pest Population - Population Invasion || Age Structured Leslie Matrix || Stage Structured Leslie Matrix || Metapopulation Dynamics -Levins Model || Interspecific Competition and Coexistence || Effect of Interspecific Competition on Species Border || Logistic Population Growth: Continuous and Discrete || Parasitoid-Host Dynamics || Conserving an Endangered Species |
| Population ecology Virtual Lab II Population ecology is the study of populations (especially population abundance) and how they change over time. Studies based on models of predation, competition as seen in interacting species is the main focus this simulation oriented lab. Lab II focuses on applied principles of population ecology for PG students. Predator - Prey Dynamics - Rats and Snakes (Lotka Volterra Simulation) || Effect of Predator Efficiency on Equilibrium Densities & Pop. Stability || Effect of Social Behavior Amongst Predator-Prey Populations || Effects of Carrying Capacity and Satiation in Predator-Prey Dynamics || Harvesting a Prey Population || Optimal Foraging with Minimal Time: A Case of Searching Predators || Optimal Foraging : Searching Predators that Maximize Energy || Optimal Pollinators || Optimal Foraging: Sit-and-wait Predators that Maximize Energy || Microparasite and Macroparasite - Host Dynamics |
| Immunology Virtual Lab I The branch of biomedicine concerned with the structure and function of the immune system, innate and acquired immunity, the bodily distinction of self from no self, and laboratory techniques involving the interaction of antigens with specific antibodies. Collection of Serum from Blood || Blood Grouping Experiment || Latex Agglutination || INDIRECT Elisa || DIRECT Elisa || SANDWICH Elisa || ELISPOT Assay || Antibody Labeling with HRP || Extraction of IgG Antibodies from Immunized Hen Egg || Isolation of lymphocytes from whole blood |
| Immunology Virtual Lab II The branch of biomedicine concerned with the structure and function of the immune system, innate and acquired immunity, the bodily distinction of self from no self, and laboratory techniques involving the interaction of antigens with specific antibodies. Ouchterlony Double Diffusion -Titration || Ouchterlony Double Diffusion - Patterns || Purification of IgG Antibodies with Ammonium Sulphate || Removal of Thymus and Spleen from Mice || Mouse Anesthesia and Blood Collection || Parenteral Injections || Purification of IgG Antibodies using Affinity Chromatography || Fluorescent Labeling of Antibodies || Fragmentation of IgG Using Papain || Fragmentation of IgG using pepsin |
| Microbiology Virtual Lab I The study of microorganisms, which are unicellular or cell-cluster microscopic organisms. This includes eukaryotes such as fungi and, protists and prokaryotes. Viruses, though not strictly classed as living organisms, are also studied. Gram Stain Technique || Aseptic Technique and the Transfer of Microorganisms || Streak Plate Method || Motility Test || Catalase and Coagulase Test || Lecithinase Test || Bacterial Growth Curve || Carbohydrate Fermentation Test || Differential and Cytological Staining Techniques || Antibiotic Susceptibility Testing || Methylene Blue Reductase Test |
| Microbiology Virtual Lab II To study the biochemical properties of microorganisms, the various techniques employed in cultivation of fungi and viruses along with the molecular level analysis of microbial genome. Voges-Proskauer Test || Triple Sugar Iron Agar || Urease Test || Litmus Milk Test || Slide Culture Technique for Fungi || Bacteriophage Plaque Assay for Phage Titer || Isolation and Identification of Auxotrophic and Drug Resistant Mutants || Isolation and Identification of Two Bacterial Unknowns || Routes of Viral Inoculation in Embryonated Eggs || 16S Ribosomal RNA Sequencing |
| Molecular Biology Virtual Lab I The study of biology at a molecular level. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry. Molecular biology chiefly concerns itself with understanding the interactions between the various systems of a cell, including the interactions between DNA, RNA and protein biosynthesis as well as learning how these interactions are regulated. Preparation of Buffer stocks (TBE,TE and TAE) || Plasmid Isolation (Mini prep) || Extraction of DNA from Fish Fins || Hot Shot Method of DNA Extraction || Agarose Gel Electrophoresis (AGE) || Restriction Digestion || Maintenance and Storage of DH5alpha E.coli cells || Preparation of Competent Cell (Calcium Chloride Treatment) || Transformation of the Host Cells || Extraction of DNA from Agarose gel |
| Molecular Biology Virtual Lab II The study of biology at a molecular level. This field overlaps with other areas of biology and chemistry, particularly genetics and biochemistry. Molecular biology chiefly concerns itself with understanding the interactions between the various systems of a cell, including the interactions between DNA, RNA and protein biosynthesis as well as learning how these interactions are regulated. Preparation of Equilibrated Phenol || Isolation of RNA || Polyacrylamide Gel Electrophoresis || Ligation ( Using T4 DNA Ligase) || Polymerase Chain Reaction (PCR) || Electroblotting || Plating of the Bacteriophage || Plasmid Curing || Extraction of Bacteriophage DNA from Large Scale Cultures Using Proteinase K and SDS || Preparation of stocks of bacteriophage lambda by plate lysis and elution |
| Cell biology Virtual Lab I Cell biology is an exciting and dynamic area that helps discover the fascinating world of cells. It includes the study of the structure and organization, growth, regulation, movements and interaction of the cells. Cell biology is closely related to other areas of biology such as genetics, molecular biology, and biochemistry. Light Microscope || Cell Organization and Sub Cellular Structure Studies (Prokaryotic and Eukaryotic) || Transmission Electron Microscopy || Isolation of Mitochondria || Isolation of Chloroplast || Isolation of Endoplasmic Reticulum || Basics of Plant Tissue Culture || Glucose Uptake Assay || Transfection || Western Blotting |
| Cell biology Virtual Lab II Cell biology is an exciting and dynamic area that helps discover the fascinating world of cells. It includes the study of the structure and organization, growth, regulation, movements and interaction of the cells. Cell biology is closely related to other areas of biology such as genetics, molecular biology, and biochemistry. Lignin Staining || Hemocytometer (Counting of Cells) || Maintenance of Mamallian Cell Lines || Cell Attachment || Cell Migration || Actin Assembly || Mitosis in Onion Root Tips || Cell Proliferation || Toxicity studies in Zebrafish || Primary Cell Culture |
| Biological Image Analysis Virtual Lab In this lab, UG/PG students will learn to use image processing techniques to analyze and quantify image data from wet lab experiments such as those in cell biology, biochemistry, molecular biology and immunology laboratories. Introduction to Biological Image Analysis || Quantification of Lignin in Tissue Sections || Analysis of Cell Morphology || Counting of Fluorescent Particles || Counting of Total Fluorescence in a Cell || Analysis on Molecular Gels: A Case Study in Polyacrylamide Gel Electrophoresis || Quantification of Stained Liver Cells || Quantification of Bacterial Colonies on an Agar Plate || Quantification of Amino Acids Present in a Mixture || Quantification of Protein Present in a Sample |
| Bioinformatics Virtual Lab I Bioinformatics is a field which using techniques of informatics to gather, store, analyse and integrate biological data. This virtual lab is an introductory course for undergraduate students and deals with the storage and retrieval of data from different biological databases like Gene, Pubmed, GEO, TAIR, Prosite etc. Retrieving sequence data from Entrez || Locating the chromosome of a Gene || Retrieve gene expression data from GEO || Retrieving articles using PubMed || Finding ORF of a Given Sequence || Retrieving structural data of a protein using PDB database || Retrieving Motif Information of a Protein Using Prosite || Retrieving Gene Information from TAIR database || Designing a primer |
| Bioinformatics Virtual Lab II This virtual laboratory is for undergraduate and postgraduate students to get a deeper understanding on the analysis of sequence data, its alignment and the evolutionary relationship. The exercises mainly deal with the different algorithms in sequence alignment and provides a computational exploration to the use of various tools used for sequence alignment. Global alignment of two sequences - Needleman-Wunsch Algorithm || Smith-Waterman Algorithm - Local Alignment of Sequences || Pairwise Sequence Alignment using BLAST || Pairwise sequence alignment using FASTA || Aligning Multiple Sequences with CLUSTAL W || Construction of Cladogram || Phylogenetic Analysis using PHYLIP - Rooted trees || Phylogenetic Analysis using PHYLIP - Unrooted trees || Genome Annotation and Multiple Sequence Allignment. |
| Bioinformatics Virtual Lab III In this virtual lab, one will study the computational analysis of proteins. This lab is targeted towards PG students with exercises that will allow one to learn visualising proteins in 3D, how to calculate distance among atoms, find active sites in protein structures and also delve into some structural analysis methods including docking and homology modeling. Combining labs 1, 2 and 3 will give an overall understanding of commonly used computational methods in bioinformatics. Visualizing the Secondary Structure of a Protein || Calculating the Distance between the Ligand and a Particular Amino acid || Finding the Active Site Pockets of a given Protein Molecule || Primary Structure Analysis of a Protein Using ProtParam || Secondary structure analysis of a protein using SOPMA || Surface Analysis of a Protein Using CASTp || Retrieving details of a drug molecule || Converting chemical file formats || Homology Modeling using Modeller || Protein- Ligand Interaction |
| Systems Biology Virtual Lab This virtual lab consist of modelling and simulation experiments for UG/PG students in bioinformatics and computational biology to understand biological processes using a systems biology approach. Mathematical modeling and simulating of Biochemical network || Import and simulate models from different databases || To Import and simulate a model from the repository || SBML-A markup language for mathematical models in systems biology using cell designer || Creating and Visualizing a Simple Network Model || Analysis of biological networks for feature detection || Integrating Biological Networks and Microarray Expression data || Analyzing the network by finding sub modules |
| Computer-Aided Drug Design Virtual Lab This lab is for PG students on the various laboratory topics in computer-aided drug design. Constructing computational model of a molecule || Introducing Hydrogen atoms to a molecule || Dihedral angle calculation of a molecule || Energy minimization of a molecule || Predict the structure of protein-Homology Modeling || Drug-Receptor Interaction || Absorption and Distribution Property Prediction in Drug Designing Process || Toxicity prediction of a Molecule |
| Ecology Virtual Lab Ecosystems are a complex and delicate balancing game.Ecosystems have an extremely complex web of cause and effect. The addition or removal of one species affects many other species with which it might compete for,or provide food. Determination of pH of Waste Water Sample || Biological Oxygen Demand || Chemical Oxygen Demand of Waste water || Nitrogen Cycle || A Brief Introduction to Species Interactions in Ecology || Bacterial Population Growth || Population Invasion - A Threat to Ecosystem || Study of Foraging of Organisms in the Ecosystem || Case Studies on Ecology |
| Bio-inspired Robotics Virtual Labs (Remote Trigger) This remote-triggerable online laboratory will teach experiments and offer to introduce biorobotics and neuronal robot techniques. The focus is on practical skills in using simple electronics to reinforce application of bio-inspired ideas. Many experiments will help working towards thesis projects. Controlling a servo motor in a bio-robotic environment (Remote Trigger) || Understanding the kinematics of a robotic upper arm (Remote Trigger) || Understanding the kinematics of a robotic upper arm - Interactive (Remote Trigger) || Light sensing process in a neural circuit (Remote Trigger) || Pattern recognition in a hardware neural network (Remote Trigger) || Mechanism behind the movement of a Walker robot with 4 neurons (Remote Trigger) || Interaction study with Neuronal Circuits || Constructing a six core brain like circuit (Remote Trigger) |
| Virtual Biophysics Lab (Remote Trigger) This lab will provide an online experience via remote equipment to study biophysics and biophysical techniques. Using a light microscope (Remote Trigger) || Observing an animal cell using a light microscope (Remote Trigger) || Study of RC Properties of Cell Membrane (Remote Trigger) || Study of Electrically excitable cells (Remote trigger) || Bursting phenomenon in biology via RC models (Remote Trigger) || Micrometry (Remote Trigger) || Multicompartmental modelling of biophysical behaviour of neurons (Remote Trigger) || Understanding Photosynthesis as a Biologically Closed Process |
| Online questionnaire for nodal centres This is a supplementary quiz series for nodal centres.All students are requested to access the quiz using their unique VALUE login ID. |
